Using deep learning to aid 3D cell detection in whole brain microscopy images
By April Cashin-Garbutt
A single mouse brain contains over 100 million neurons. Analysing only a small percentage of these cells manually is a daunting task and one that can take many days, if not weeks, to complete. Researchers in the Margrie lab at the Sainsbury Wellcome Centre have developed an algorithm that can accurately detect cells in whole mouse brain images in a mere fraction of the time it takes to accomplish manually.
“Due to advances in imaging technology, we can now acquire whole mouse brain images in half a day and we wanted to be able to analyse the data in a similar timeframe. That’s why we developed an algorithm that can quickly and accurately complete 3D cell detection in whole mouse brain images using standard computer hardware” said Adam Tyson, Research Fellow in the Margrie Lab at the Sainsbury Wellcome Centre.
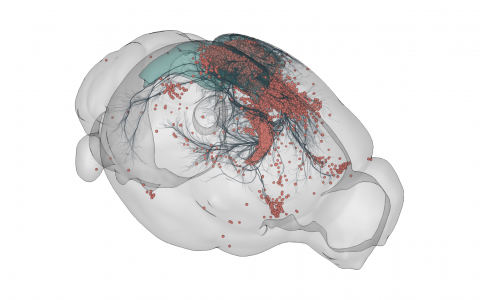
3D rendering of labelled mouse brain cells detected by cellfinder (coral), visualised alongside anatomical tracing data (blue) from the Allen Mouse Brain Connectivity Atlas
While deep learning based approaches are revolutionising image analysis, they typically require a lot of computer resources and can be relatively slow to process large datasets. Adam Tyson, Christian Niedworok and Charly Rousseau from the Margrie lab created a two-step process to overcome this challenge for whole mouse brain images. The cell-detection algorithm they developed combines both classical imaging and deep learning. First, a classical imaging step detects which parts of the image need to be looked at again. This subset of the image is then fed into a more sophisticated deep learning algorithm to complete the 3D cell detection.
This novel approach means that the complicated method is only used for the parts of the data that are difficult to analyse. In the same way that we would only use a calculator for the parts of a sum that are hard to do in our head, deep learning is only used for the parts of the image where the cells need to be detected.
Unlike most deep learning based tools, which require a reasonable amount of programming knowledge, the cell detection algorithm that Adam and team have created includes a plug-in for napari. This means that neuroscientists can easily apply the algorithm to their data using an intuitive graphical user interface.
The algorithm is a component of cellfinder, a software package that, in addition to cell detection, assigns cells to brain regions. The cellfinder software was developed as part of the BrainGlobe Initiative, a multi-national consortium of neuroscientists and developers working together to generate open-source, interoperable and easy-to-use Python-based tools for computational neuroanatomy.
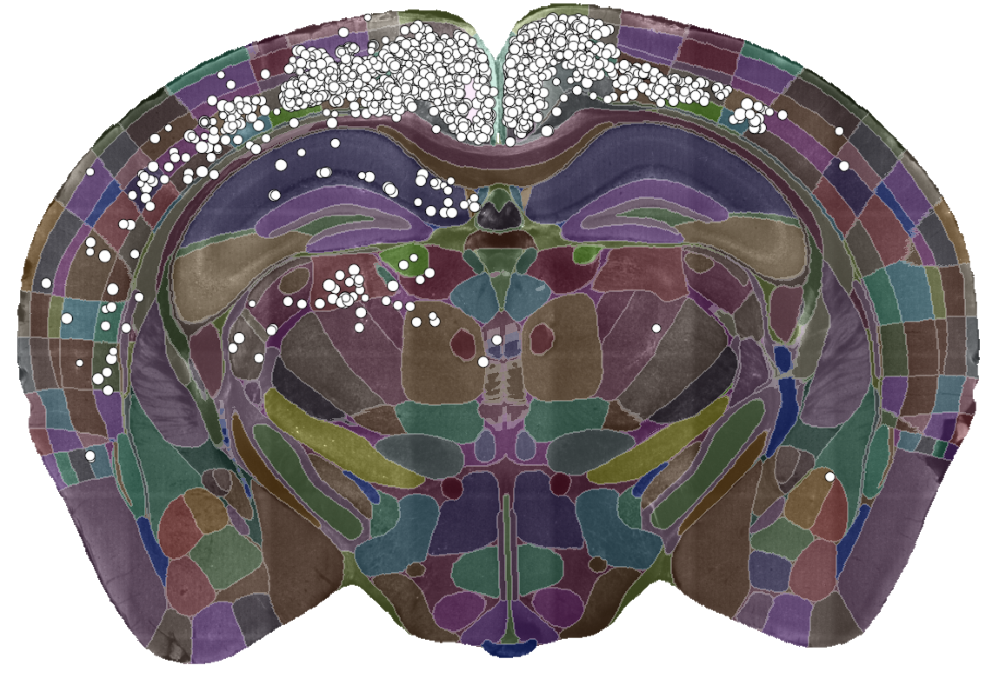
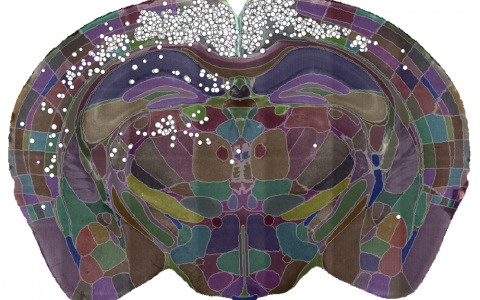
A single slice of a 3D brain image showing the detected cells (white dots) and the brain divided into regions (coloured sections). Data courtesy of Dr Sepiedeh Keshavarzi and Dr Chryssanthi Tsitoura.
To demonstrate the power of the cell-detection algorithm, Adam and team used it to analyse tracing data looking at cells that provide input into primary visual cortex (V1). In a new paper in PLOS Computational Biology they show how the algorithm can reproduce known findings of which areas project to V1. They also found very high correlation with expert manual counts. The algorithm, however, was much faster. While experts would take a day or two to complete a manual count, the algorithm took about 90 minutes on a normal computer. In addition, the computer was completely unattended during this time, which opens up the possibility of carrying out such experiments much more routinely.
The algorithm also opens up the method to more labs as it doesn’t require any dedicated high performance computing. In addition, it is a reproducible method, which is good for the neuroscience community. The team were surprised to find that the results of the algorithm can be improved with relatively little training data. When they trained the algorithm using data from one microscope and then applied it to another one, only minimal retraining was needed to make it perform well on the new data.
The next steps for the team are to extend the algorithm so that it can achieve cell-type classification. This would mean that the algorithm could detect not only that a cell was present, but whether it was cell type A or B or an artefact for example.
Read the paper in PLOS Computational Biology: A deep learning algorithm for 3D cell detection in whole mouse brain image datasets