The BrainGlobe Initiative
The BrainGlobe Initiative (BGI) was created by neuroscientists at the Sainsbury Wellcome Centre, UCL and the Max Planck Institute for Neurobiology (now at the Technical University of Munich) and is now a multi-national consortium of neuroscientists and developers from Europe, UK and USA (including Princeton University, the Kavli Institute for Systems Neuroscience, Tel Aviv University and the Francis Crick Institute). The central goal of the BGI is to establish a critical mass of scientists and programmers that can work together to generate open-source, interoperable and easy to use Python-based tools for computational neuroanatomy.
As image datasets become more complex and very large 3D datasets become easier to acquire (e.g. the mesoSPIM project) the neuroscience community requires a central platform for computational neuroanatomy that is easy to use and is well integrated with other related efforts (e.g. napari) and modern data analysis frameworks. BGI aims to be such a platform: the development of BGI was aimed at addressing the main shortcomings of most currently available software for neuroanatomy. Most software tools to date are designed to work with a single brain atlas or model species, requiring that additional, often duplicated, effort be spent into developing similar software for other model species. Furthermore these software are generally not written in Python, which is becoming the programming language of choice for the neuroscientific community due to other open-source efforts, such as DeepLabCut.
The BGI aims to address these issues by developing a suite of software tools, written in Python, for computational neuroanatomy. BGI software is fully integrated facilitating the development of complex data analysis pipelines. In addition, BGI software can work with any atlas and model species by utilising BGI’s own AtlasAPI software (Claudi, Petrucco & Tyson 2021). This API can be re-used allowing others to develop software compatible with all existing BrainGlobe tools. BGI’s software was designed to be easy to use by researchers and extensive online documentation and examples are provided to help new users learn how to use this software.
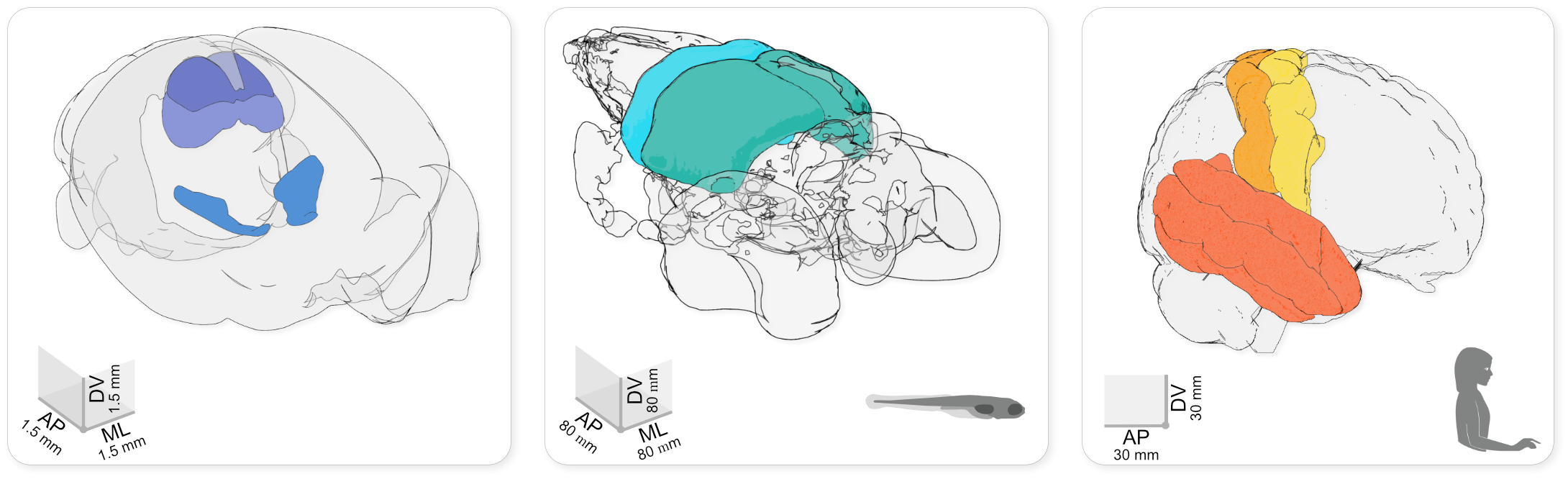
Using brainrender to visualise brain atlas data in mouse, larval zebrafish and human. The brainrender GUI. Mouse, human, and zebrafish larvae drawings from scidraw.io (doi.org/10.5281/zenodo.3925991, doi.org/10.5281/zenodo.3926189, doi.org/10.5281/zenodo.3926123).
The BGI focuses on sustainability of its software and aims to achieve this through collaboration with neuroscientists (>20 contributors to date) and with the scientific python ecosystem. All tools developed by the BGI are open source meaning that they can be used, and adapted by researchers around the world for free. These include brainreg and brainglobe-segmentation (Tyson, Vélez-Fort & Rousseau et al. 2022) for the delineation of structures in whole-brain microscopy images and cellfinder (Tyson, Rousseau & Niedworok et al. 2021), a novel approach for cell detection in these images. The BGI have also recently published brainrender (Claudi et al. 2021), software for the visualisation of multimodal neuroanatomical data in a common coordinate space (e.g. that from cellfinder or brainreg).
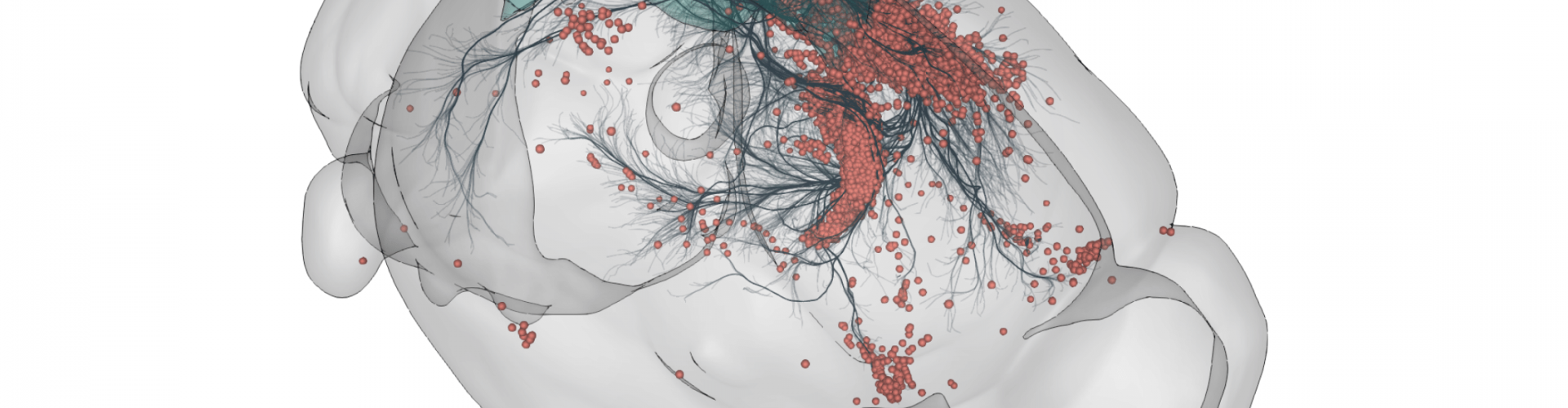
3D rendering of labelled mouse brain cells detected by cellfinder (coral), visualised alongside anatomical tracing data (blue) from the Allen Mouse Brain Connectivity Atlas
The BGI always welcomes new users and contributors, regardless of their programming experience. These tools are built by scientists, for scientists and any input (including bug reports and features requests) is incredibly valuable. To read more about these tools, see the BrainGlobe website, or feel free to ask us a question on the image.sc forum. More details about individual software tools are available on their respective GitHub repositories:
Publications
Niedworok, C., Brown, A., Jorge Cardoso, M. et al. (2016) aMAP is a validated pipeline for registration and segmentation of high-resolution mouse brain data. Nat Commun 7, 11879 https://doi.org/10.1038/ncomms11879
Claudi, F.*, Petrucco, L.*, Tyson, A.L.*, Branco, T., Margrie, T.W., Portugues, R. (2020) BrainGlobe Atlas API: a common interface for neuroanatomical atlases. J. Open Source Software. 5, 2668 https://doi.org/10.21105/joss.02668
Claudi F., Tyson A.L., Petrucco L., Margrie T.W., Portugues R., Branco T. (2021) Visualizing anatomically registered data with brainrender. eLife 10:e65751 https://doi.org/10.7554/eLife.65751
Tyson, A.L.*, Rousseau, C.V.*, Niedworok, C.J.*, Keshavarzi, S., Tsitoura, C., Cossell, L., Strom, M., Margrie, T.W. (2021) A deep learning algorithm for 3D cell detection in whole mouse brain image datasets. PloS Computational Bio 17(5): e1009074 https://doi.org/10.1371/journal.pcbi.1009074
Tyson, A.L.*, Vélez-Fort, M.*, Rousseau, C.V.*, Cossell, L., Tsitoura, C., Lenzi, S. C., Obenhaus, H.A., Claudi, F., Branco, T., Margrie, T.W. (2022) Accurate determination of marker location within whole-brain microscopy images. Scientific Reports 12, 867 https://doi.org/10.1038/s41598-021-04676-9