Chan Zuckerberg Initiative awards two grants to BrainGlobe
Computational neuroanatomy applications awarded napari Plugin Accelerator Grants
BrainGlobe, an open-source, community-driven platform for computational neuroanatomy, has today been awarded two grants from the Chan Zuckerberg Initiative (CZI).
“One of the missions of the CZI is to accelerate progress in science by supporting the development of transformative technologies. BrainGlobe fits this vision by providing a central platform for computational neuroanatomy that is easy-to-use and well-integrated with other efforts including napari,” said Troy Margrie, Associate Director of the Sainsbury Wellcome Centre and co-PI on the CZI grant awarded to BrainGlobe.
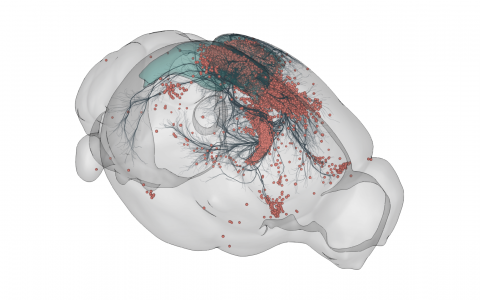
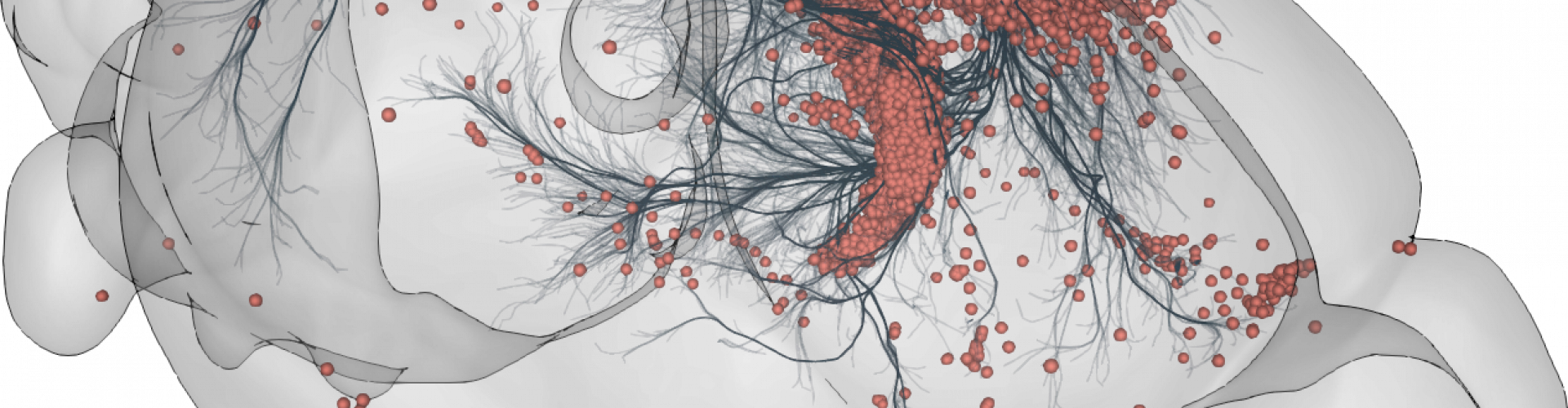
3D rendering of labelled mouse brain cells detected by cellfinder (coral), visualised alongside anatomical tracing data (blue) from the Allen Mouse Brain Connectivity Atlas
The two grants awarded to BrainGlobe by CZI were in response to their Request for Applications for napari Plugin Accelerator Grants. This funding call was launched to support plugin projects for the napari image analysis platform, with the aim of generating an extensible plugin ecosystem to provide easy access to reproducible and quantitative bioimage analysis.
“Although very sophisticated, napari itself has very limited functionality for biologists and so the aim is to allow people to develop plugins for biological applications, including both general image processing algorithms and more specific applications,” said Adam Tyson, former member of The Margrie Lab and now the Scientific Software Lead at the Institute of Cancer Research and co-PI on the CZI napari Plugin Accelerator Grant awarded to BrainGlobe.
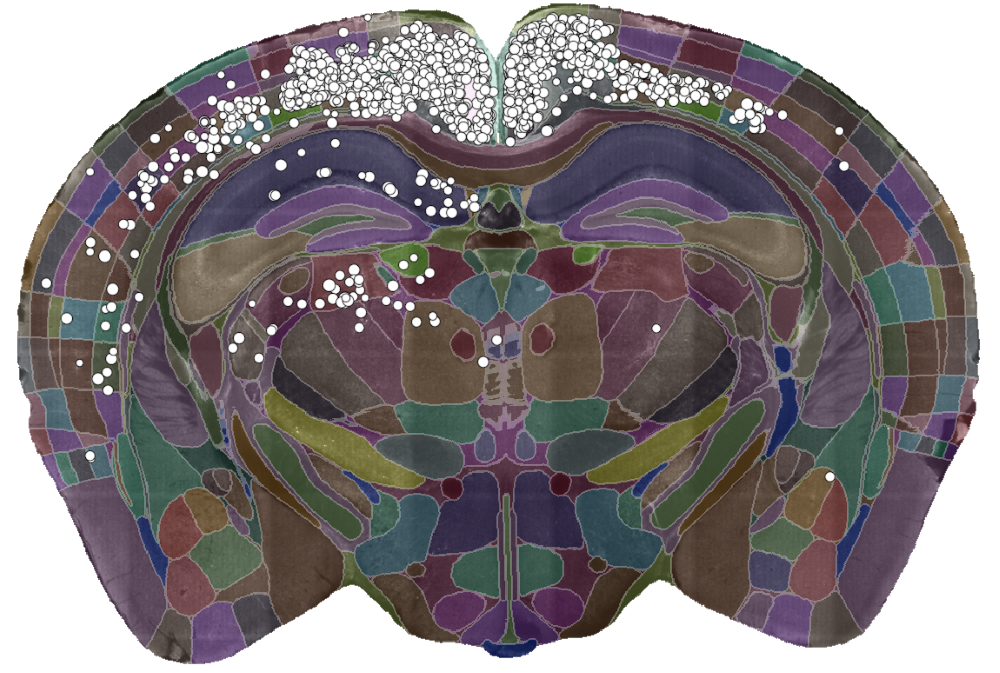
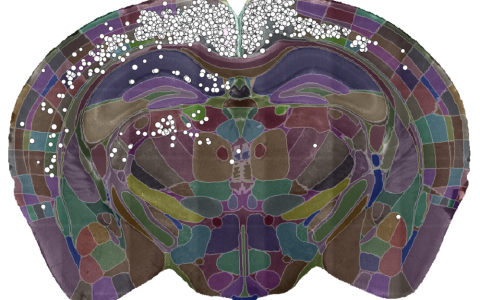
A single slice of a 3D brain image showing the detected cells (white dots) and the brain divided into regions (coloured sections). Data courtesy of Dr Sepiedeh Keshavarzi and Dr Chryssanthi Tsitoura.
The first of the two grants awarded to the team was to help maintain and develop existing software called cellfinder, which was developed by BrainGlobe for the analysis of whole-brain imaging data. The cellfinder software provides a novel approach for cell-detection in whole-brain images, thereby bringing terabyte-scale 3D cell-detection to napari. The second grant was awarded to develop a new meta-plugin to bring together multiple different software plugins within the BrainGlobe initiative (BGI) so that they can all work in an interoperative way within a single environment. Both grants will allow the BrainGlobe plugins to be developed and maintained for the community, while also making them more robust due to the funding of specific development time. Through collaboration with UCL Scientific Computing this work aims to be completed by July 2022.
In addition to the grants themselves, the launch of the program will kick-off with a virtual event gathering together all the napari Plugin Accelerator grantees in December to celebrate the awards and connect with each other and CZI. This will help build connections between the grantees and further develop the napari community.
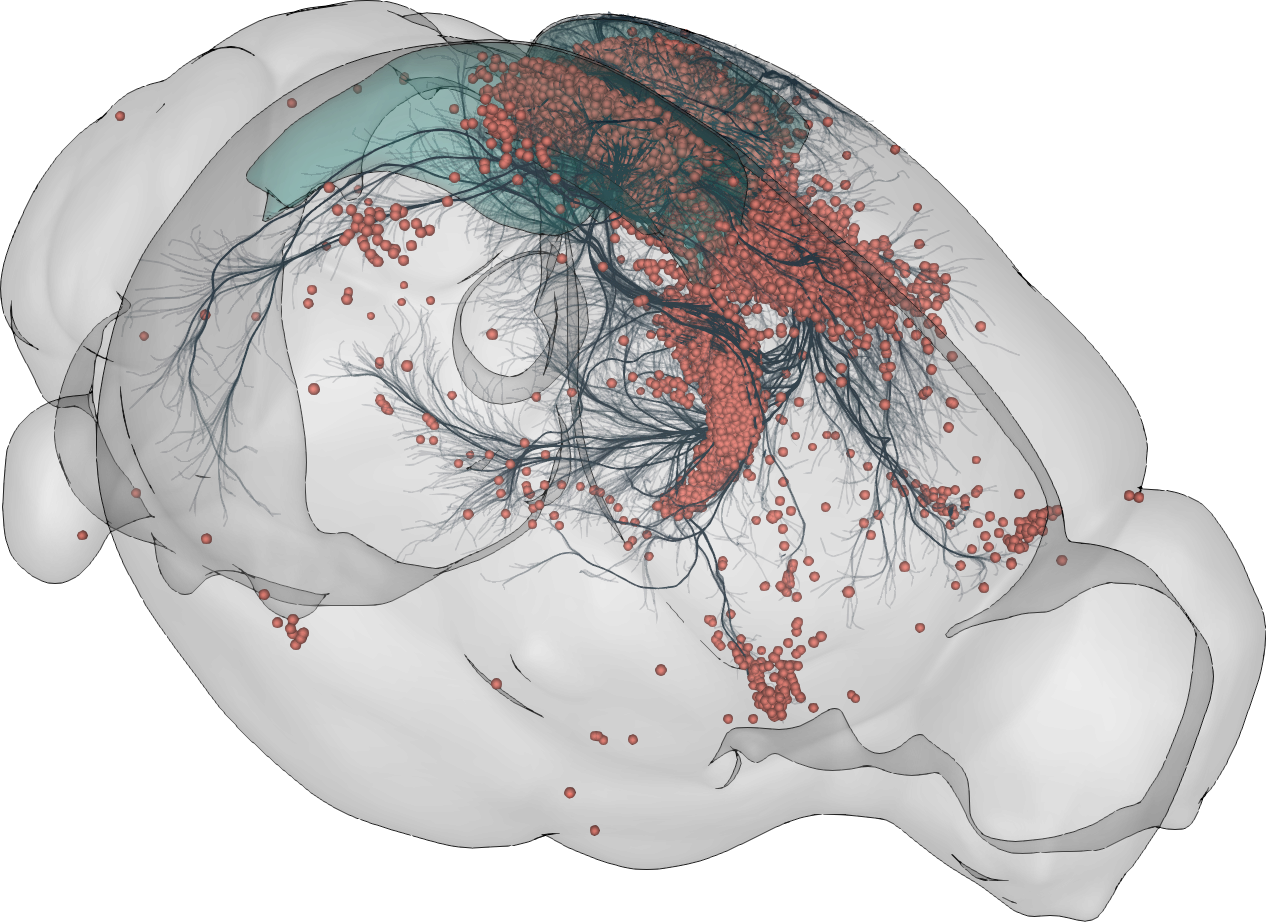
3D rendering of labelled mouse brain cells by cellfinder (coral), visualised alongside anatomical tracing data (blue) from the Allen Mouse Brain Connectivity Atlas.
“Although the grantees may be working in different areas, fundamentally a lot of image analysis ends up falling down to the same methods. Bringing together the napari Plugin Accelerator grantees will help to develop a community of people who are writing software for the same platform, which will hopefully lead to fruitful collaborations and also make sure we avoid duplication of efforts,” commented Adam Tyson. “It’s great to see that all of the hard work started almost ten years ago (at NIMR) by various people including Christian Niedworok and Charly Rousseau to develop many of these validated analytical pipelines for open access (that now sit under the BGI banner), come to fruition. Their high download numbers probably reflect the desperate need for these tools that are already contributing to impactful science,” Troy Margrie commented.
Media contact:
For more information or to speak to the researchers involved, please contact:
- April Cashin-Garbutt
Head of Research Communications and Engagement
E: a.cashin-garbutt@ucl.ac.uk T: +44 (0)20 3108 8028
About the BrainGlobe Initiative
The BrainGlobe Initiative (BGI) was created by neuroscientists at the Sainsbury Wellcome Centre, UCL and the Max Planck Institute for Neurobiology (now at the Technical University of Munich) and is now a multi-national consortium of neuroscientists and developers from Europe, UK and USA. The central goal of the BGI is to establish a critical mass of scientists and programmers that can work together to generate open-source, interoperable and easy to use Python-based tools for computational neuroanatomy. For more information, please visit: https://www.sainsburywellcome.org/web/content/brainglobe-initiative
About the Sainsbury Wellcome Centre
The Sainsbury Wellcome Centre (SWC) brings together world-leading neuroscientists to generate theories about how neural circuits in the brain give rise to the fundamental processes underpinning behaviour, including perception, memory, expectation, decisions, cognition, volition and action. Funded by the Gatsby Charitable Foundation and Wellcome, SWC is located within UCL and is closely associated with the Faculties of Life Sciences and Brain Sciences.
About the Chan Zuckerberg Initiative
The Chan Zuckerberg Initiative was founded in 2015 to help solve some of society’s toughest challenges — from eradicating disease and improving education, to addressing the needs of our local communities. Our mission is to build a more inclusive, just, and healthy future for everyone. For more information, please visit: www.chanzuckerberg.com